Fibermagic Tutorial¶
Fibermagic is an open-source software package for the analysis of photometry data. It is written in Python and operates on pandas and numpy dataframes.
Fiber Photometry is a novel technique to capture neural activity in-vivo with a subsecond temporal precision. Genetically encoded fluorescent sensors are expressed by either transfection with an (AAV)-virus or transgenic expression. Strenght of fluorescence is dependent on the concentration of the neurotransmitter the sensor detects. Using an optic glas fiber, it is possible to image neural dynamics and transmitter release.

graphik source: https://en.wikipedia.org/wiki/Fiber_photometry
Fiber Photometry using Neurophotometrics devices¶
There are a bunch of different photometry systems out there like TDT systems, Neurophotometrics or open-hardware approaches. In this tutorial, we will use data from Neurophotometrics as an example.

Multiple Colors¶
With Neurophotometrics (NPM), it is possible to capture data from two fluorescent sensors simultaneously - if they emit light in different wave lengths. NPM can measure light of two different color spectra: Red and Green. Using this technology, it is possible, to e.g. express a green calcium sensor (e.g. GCaMP6f) and a red dopamine sensor (e.g. Rdlight1).
 grafik source: https://www.researchgate.net/publication/264796291_Bright_and_fast_multi-colored_voltage_reporters_via_electrochromic_FRET/figures?lo=1
grafik source: https://www.researchgate.net/publication/264796291_Bright_and_fast_multi-colored_voltage_reporters_via_electrochromic_FRET/figures?lo=1
Multiple Mice¶
NPM can record data from multiple mice at once - delivered through a single path chord that splits into several small cables that can be attached individually to a single mouse.
The end of the patch chord projects to a photosensor which captures the light. Here’s how it looks like:

Before starting the recording, the researcher defines a region of interest for each cable and color. Then, NPM captures the light intensity for each region of interest seperately. All data streams are written into one single csv file per recording.
Investigating a single raw data file¶
Let’s have a look into an arbitrary recording file and try to understand the data.
[1]:
!pip install fibermagic
import pandas as pd
from fibermagic.utils.download_dataset import download
download('tutorial')
Requirement already satisfied: fibermagic in c:\users\georg\git\fibermagic (0.1.4)
Requirement already satisfied: numpy in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from fibermagic) (1.21.4)
Requirement already satisfied: pandas in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from fibermagic) (1.3.4)
Requirement already satisfied: matplotlib in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from fibermagic) (3.5.1)
Requirement already satisfied: scipy in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from fibermagic) (1.7.2)
Requirement already satisfied: scikit-learn in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from fibermagic) (1.0.1)
Requirement already satisfied: plotly in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from fibermagic) (5.4.0)
Requirement already satisfied: dash in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from fibermagic) (2.0.0)
Requirement already satisfied: Flask>=1.0.4 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from dash->fibermagic) (2.0.2)
Requirement already satisfied: dash-table==5.0.0 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from dash->fibermagic) (5.0.0)
Requirement already satisfied: dash-core-components==2.0.0 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from dash->fibermagic) (2.0.0)
Requirement already satisfied: dash-html-components==2.0.0 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from dash->fibermagic) (2.0.0)
Requirement already satisfied: flask-compress in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from dash->fibermagic) (1.10.1)
Requirement already satisfied: tenacity>=6.2.0 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from plotly->fibermagic) (8.0.1)
Requirement already satisfied: six in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from plotly->fibermagic) (1.16.0)
Requirement already satisfied: packaging>=20.0 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from matplotlib->fibermagic) (21.3)
Requirement already satisfied: pyparsing>=2.2.1 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from matplotlib->fibermagic) (3.0.6)
Requirement already satisfied: pillow>=6.2.0 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from matplotlib->fibermagic) (8.4.0)
Requirement already satisfied: fonttools>=4.22.0 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from matplotlib->fibermagic) (4.28.2)
Requirement already satisfied: python-dateutil>=2.7 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from matplotlib->fibermagic) (2.8.2)
Requirement already satisfied: cycler>=0.10 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from matplotlib->fibermagic) (0.11.0)
Requirement already satisfied: kiwisolver>=1.0.1 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from matplotlib->fibermagic) (1.3.2)
Requirement already satisfied: pytz>=2017.3 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from pandas->fibermagic) (2021.3)
Requirement already satisfied: joblib>=0.11 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from scikit-learn->fibermagic) (1.1.0)
Requirement already satisfied: threadpoolctl>=2.0.0 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from scikit-learn->fibermagic) (3.0.0)
Requirement already satisfied: Werkzeug>=2.0 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from Flask>=1.0.4->dash->fibermagic) (2.0.2)
Requirement already satisfied: click>=7.1.2 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from Flask>=1.0.4->dash->fibermagic) (8.0.3)
Requirement already satisfied: itsdangerous>=2.0 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from Flask>=1.0.4->dash->fibermagic) (2.0.1)
Requirement already satisfied: Jinja2>=3.0 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from Flask>=1.0.4->dash->fibermagic) (3.0.3)
Requirement already satisfied: brotli in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from flask-compress->dash->fibermagic) (1.0.9)
Requirement already satisfied: colorama in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from click>=7.1.2->Flask>=1.0.4->dash->fibermagic) (0.4.4)
Requirement already satisfied: MarkupSafe>=2.0 in c:\users\georg\anaconda3\envs\fibermagic\lib\site-packages (from Jinja2>=3.0->Flask>=1.0.4->dash->fibermagic) (2.0.1)
Downloading started
Downloading Completed, Extracting...
[2]:
df = pd.read_csv('rawdata_demo.csv')
df.head(10)
[2]:
| FrameCounter | Timestamp | Flags | Region0R | Region1G | Region2R | Region3G | Region4R | Region5G | Region6R | Region7G | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 2955.990560 | 16 | 0.003922 | 0.003922 | 0.003922 | 0.003922 | 0.003922 | 0.003922 | 0.003922 | 0.003922 |
| 1 | 1 | 2956.003840 | 18 | 0.004341 | 0.007010 | 0.005176 | 0.008868 | 0.004264 | 0.006001 | 0.005350 | 0.007242 |
| 2 | 2 | 2956.017184 | 20 | 0.020586 | 0.350305 | 0.020055 | 0.310345 | 0.018077 | 0.380686 | 0.011893 | 0.189797 |
| 3 | 3 | 2956.030528 | 17 | 0.018014 | 0.015875 | 0.019811 | 0.017195 | 0.019681 | 0.024707 | 0.015898 | 0.014165 |
| 4 | 4 | 2956.043840 | 18 | 0.003923 | 0.004603 | 0.003935 | 0.007462 | 0.003922 | 0.004054 | 0.003966 | 0.004644 |
| 5 | 5 | 2956.057184 | 20 | 0.018542 | 0.344338 | 0.018014 | 0.304967 | 0.015870 | 0.374825 | 0.009874 | 0.185943 |
| 6 | 6 | 2956.070528 | 17 | 0.017936 | 0.015720 | 0.019849 | 0.017191 | 0.019787 | 0.024617 | 0.015881 | 0.014059 |
| 7 | 7 | 2956.083840 | 18 | 0.004350 | 0.007031 | 0.005192 | 0.008787 | 0.004257 | 0.005926 | 0.005291 | 0.007185 |
| 8 | 8 | 2956.097184 | 20 | 0.020549 | 0.345224 | 0.019962 | 0.305642 | 0.017845 | 0.375177 | 0.011721 | 0.187082 |
| 9 | 9 | 2956.110528 | 17 | 0.017933 | 0.015715 | 0.019841 | 0.017110 | 0.019732 | 0.024621 | 0.015832 | 0.014013 |
Each Region of Interest is represented as one column.
A simple frame counter and a timestamp indicate when a measurement happened.
The Flags column encodes different things: Which LED was on at the time of measurement, which input and/or output channel was on or off and whether the laser for optogenetic stimulation was on. NPM encodes all this information into a single number using a dual encoding.
Because the emission spectra of fluorescent proteins are partly overlapping, it is not possible to measure both, the red and green sensor, at the same time. Instead, only one LED is on at a time. That way, we get presice measurements.
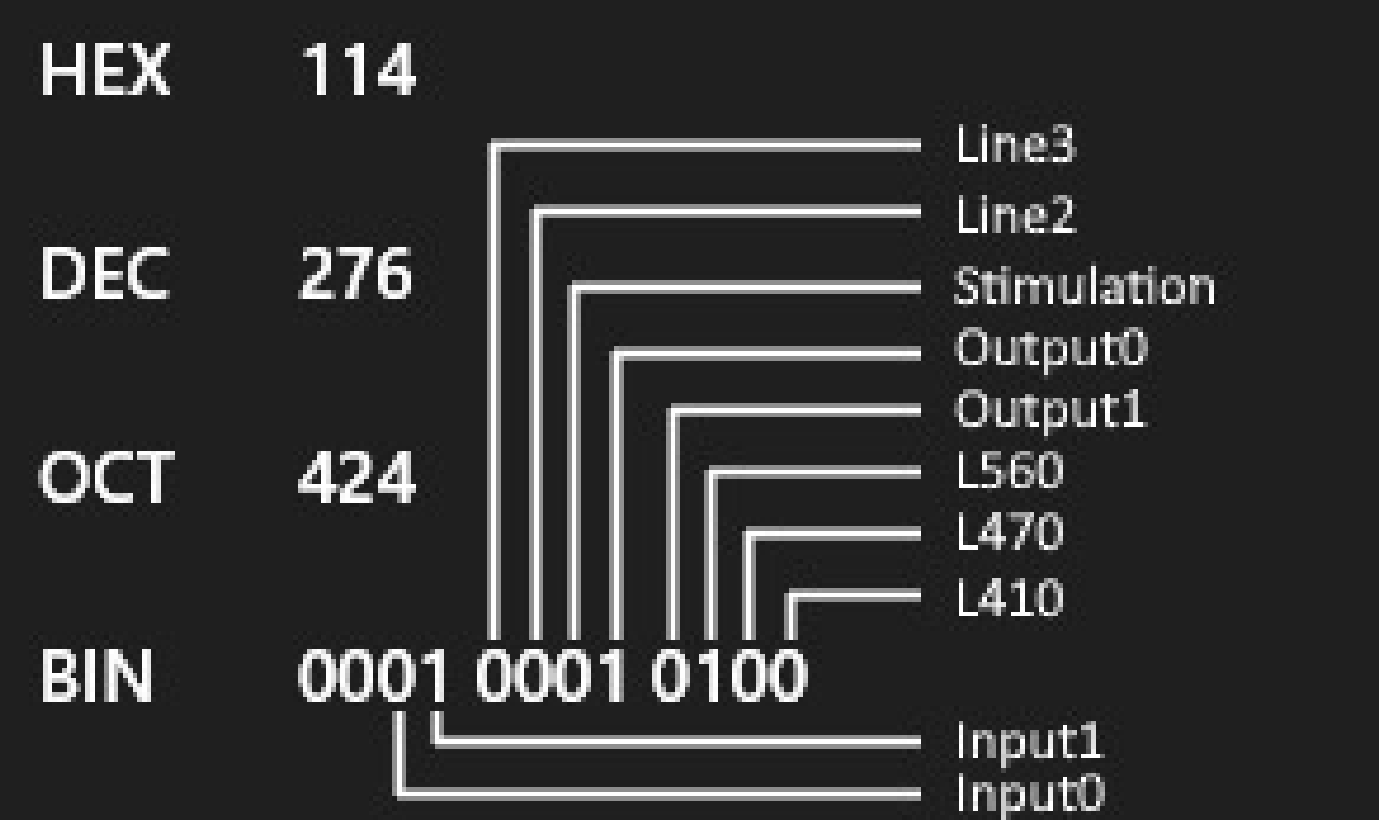
We can use fibermagic to decode which LEDs were on at what measurement from the Flags column.
[3]:
from fibermagic.IO.NeurophotometricsIO import extract_leds
[4]:
if 'Flags' in df.columns: # legacy fix: Flags were renamed to LedState
df = df.rename(columns={'Flags': 'LedState'})
df = extract_leds(df).dropna()
df.head(10)
[4]:
| FrameCounter | Timestamp | LedState | Region0R | Region1G | Region2R | Region3G | Region4R | Region5G | Region6R | Region7G | wave_len | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 2956.003840 | 18 | 0.004341 | 0.007010 | 0.005176 | 0.008868 | 0.004264 | 0.006001 | 0.005350 | 0.007242 | 470.0 |
| 2 | 2 | 2956.017184 | 20 | 0.020586 | 0.350305 | 0.020055 | 0.310345 | 0.018077 | 0.380686 | 0.011893 | 0.189797 | 560.0 |
| 3 | 3 | 2956.030528 | 17 | 0.018014 | 0.015875 | 0.019811 | 0.017195 | 0.019681 | 0.024707 | 0.015898 | 0.014165 | 410.0 |
| 4 | 4 | 2956.043840 | 18 | 0.003923 | 0.004603 | 0.003935 | 0.007462 | 0.003922 | 0.004054 | 0.003966 | 0.004644 | 470.0 |
| 5 | 5 | 2956.057184 | 20 | 0.018542 | 0.344338 | 0.018014 | 0.304967 | 0.015870 | 0.374825 | 0.009874 | 0.185943 | 560.0 |
| 6 | 6 | 2956.070528 | 17 | 0.017936 | 0.015720 | 0.019849 | 0.017191 | 0.019787 | 0.024617 | 0.015881 | 0.014059 | 410.0 |
| 7 | 7 | 2956.083840 | 18 | 0.004350 | 0.007031 | 0.005192 | 0.008787 | 0.004257 | 0.005926 | 0.005291 | 0.007185 | 470.0 |
| 8 | 8 | 2956.097184 | 20 | 0.020549 | 0.345224 | 0.019962 | 0.305642 | 0.017845 | 0.375177 | 0.011721 | 0.187082 | 560.0 |
| 9 | 9 | 2956.110528 | 17 | 0.017933 | 0.015715 | 0.019841 | 0.017110 | 0.019732 | 0.024621 | 0.015832 | 0.014013 | 410.0 |
| 10 | 10 | 2956.123840 | 18 | 0.004425 | 0.007011 | 0.005275 | 0.008732 | 0.004260 | 0.005959 | 0.005242 | 0.007178 | 470.0 |
Now, we have all we need to plot the full trace of a single sensor of one mouse and inspect the raw data values. In this recording, Rdlight1 and GCaMP6f were expressed. Rdlight1 is excited by the 560 nm LED and emits light in the red color spectrum. GCaMP6f is excited by the 470 nm LED and emits light in the green spectrum. Now, let’s spike into the raw data of Rdlight1 of a single mouse.
We use “plotly” for plotting here. Plotly is a plotting library for Python and other programming languages. It is interactive, which means we can zoom in and out and scroll through the data, which is exactly what we want to do at the moment.
[5]:
import plotly.express as px
[6]:
region0R_red = df[df.wave_len == 560][['FrameCounter', 'Region0R']]
px.line(region0R_red, x='FrameCounter', y='Region0R')
We can clearly see huge transients from dopamine activity in the raw data. Congratulations!
However, there are several issues with the data. Fiber Photometry is not measureing the difference between day and night. Changes in fluorescence are usually extremely small and very hard to measure. We basically measure light intensity with a super high precision. However, this means that the photometry system will also pick up every disturbance, even if very small.
Examples of unwanted noise are: * Photobleaching of the Patch Chord * Photobleaching caused by LED heating * Photobleaching caused by destroyment of sensors * Motion artifacts * Loose or poorly attached cables
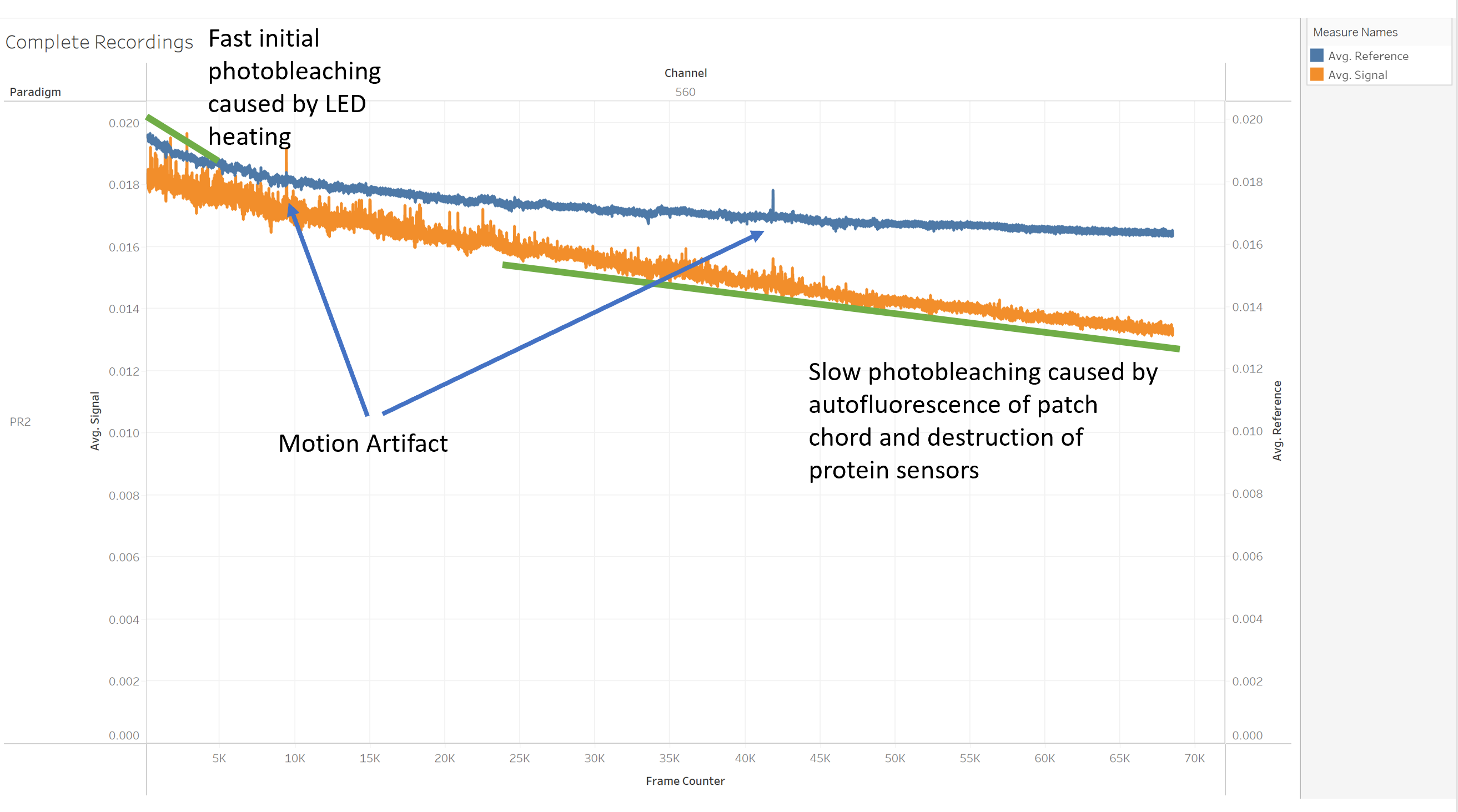
Demodulation of Fiber Photometry Data¶
As we saw, the raw data is useless without further processing. There are a variety of different of different methods to remove photobleaching and motion artifacts, here are a few examples:
Correction of Photobleaching:¶
High-Pass Filtering: Photobleaching usually occurs at a very slow timescale. By applying a simple high-pass filter, e.g. a butterworth, it is possible to remove the gross artifacts of photobleaching, but it removes also slow changes in neurotransmitter concentration
Biexponential decay: Photobleaching can be estimated by a decreasing exponential function. However, as it happens on two different timescales (e.g. fast LED-heating-based photobleaching and slow patch chord photobleaching), we need a biexponential decay that can be regressed to the data and then subtracted.
airPLS: Adaptive iteratively reweighted penalized least squares. A more advanced method to remove artifacts. For more info, please see the paper: DOI: 10.1039/b922045c
Correction of Motion Artifacts:¶
Genetically encoded Neurotransmitter indicators have usually a useful attribute: If stimulated at around 410 nm (instead of e.g. 470 or 560), the excitation will be the same independent of the neurotransmitter concentration. Stimulating a this wave length is called recording an “isobestic”. The isobestic is usefull to correct motion artifacts.
If a transient is caused by neural activity, it should be detectable in the signal channel, but if it is caused by motion, it should be detectable in both, the isobestic and signal.
We can remove motion artifacts if we fit the isobestic to the signal and subtract.
Demodulate using airPLS¶
We can use fibermagic to calculate z-scores of dF/F using the airPLS algorithm. However, before we do that, we need to bring the dataframe into long-format.
[7]:
NPM_RED = 560
NPM_GREEN = 470
NPM_ISO = 410
# dirty hack to come around dropped frames until we find better solution -
# it makes about 0.16 s difference
df.FrameCounter = df.index // len(df.wave_len.unique())
df = df.set_index('FrameCounter')
regions = [column for column in df.columns if 'Region' in column]
dfs = list()
for region in regions:
channel = NPM_GREEN if 'G' in region else NPM_RED
sdf = pd.DataFrame(data={
'Region': region,
'Channel': channel,
'Signal': df[region][df.wave_len == channel],
'Reference': df[region][df.wave_len == NPM_ISO]
}
)
dfs.append(sdf)
dfs = pd.concat(dfs).reset_index().set_index(['Region', 'Channel', 'FrameCounter'])
dfs
[7]:
| Signal | Reference | |||
|---|---|---|---|---|
| Region | Channel | FrameCounter | ||
| Region0R | 560 | 0 | 0.020586 | NaN |
| 1 | 0.018542 | 0.018014 | ||
| 2 | 0.020549 | 0.017936 | ||
| 3 | 0.020701 | 0.017933 | ||
| 4 | 0.021635 | 0.018000 | ||
| ... | ... | ... | ... | ... |
| Region7G | 470 | 68674 | 0.005743 | 0.010064 |
| 68675 | 0.005734 | 0.010049 | ||
| 68676 | 0.005767 | 0.010081 | ||
| 68677 | 0.005793 | 0.010060 | ||
| 68678 | NaN | 0.010077 |
549432 rows × 2 columns
[8]:
from fibermagic.core.demodulate import add_zdFF
dfs = add_zdFF(dfs, method='airPLS', remove=250)
[9]:
dfs
[9]:
| FrameCounter | Signal | Reference | zdFF (airPLS) | ||
|---|---|---|---|---|---|
| Region | Channel | ||||
| Region0R | 560 | 251 | 0.020384 | 0.017583 | -1.751040 |
| 560 | 252 | 0.020382 | 0.017586 | -1.751065 | |
| 560 | 253 | 0.020450 | 0.017540 | -1.715426 | |
| 560 | 254 | 0.020423 | 0.017533 | -1.596002 | |
| 560 | 255 | 0.020440 | 0.017572 | -1.544472 | |
| ... | ... | ... | ... | ... | ... |
| Region7G | 470 | 68423 | 0.005653 | 0.010038 | -1.173983 |
| 470 | 68424 | 0.005743 | 0.010010 | -0.189733 | |
| 470 | 68425 | 0.005760 | 0.010022 | 0.253415 | |
| 470 | 68426 | 0.005812 | 0.009984 | 0.270365 | |
| 470 | 68427 | 0.005806 | 0.009965 | 0.285169 |
545416 rows × 4 columns
[10]:
fig = px.line(dfs.reset_index(), x='FrameCounter', y='zdFF (airPLS)', facet_row='Region', height=1000)
fig.for_each_annotation(lambda a: a.update(text=a.text.split("=")[-1]))
fig